Fast spatial indexing with Priority R-Tree for Python. Efficiently query 2D/3D/4D bounding boxes with C++ performance.
pip install python-prtreeimport numpy as np
from python_prtree import PRTree2D
# Create rectangles: [xmin, ymin, xmax, ymax]
rects = np.array([
[0.0, 0.0, 1.0, 0.5], # Rectangle 1
[1.0, 1.5, 1.2, 3.0], # Rectangle 2
])
indices = np.array([1, 2])
# Build the tree
tree = PRTree2D(indices, rects)
# Query: find rectangles overlapping with [0.5, 0.2, 0.6, 0.3]
result = tree.query([0.5, 0.2, 0.6, 0.3])
print(result) # [1]
# Batch query (faster for multiple queries)
queries = np.array([
[0.5, 0.2, 0.6, 0.3],
[0.8, 0.5, 1.5, 3.5],
])
results = tree.batch_query(queries)
print(results) # [[1], [1, 2]]- Construction: Create from numpy arrays (2D, 3D, or 4D)
- Query: Find overlapping bounding boxes
- Batch Query: Parallel queries for high performance
- Insert/Erase: Dynamic updates (optimized for mostly static data)
- Query Intersections: Find all pairs of intersecting boxes
- Save/Load: Serialize tree to disk
from python_prtree import PRTree2D, PRTree3D, PRTree4D
tree2d = PRTree2D(indices, boxes_2d) # [xmin, ymin, xmax, ymax]
tree3d = PRTree3D(indices, boxes_3d) # [xmin, ymin, zmin, xmax, ymax, zmax]
tree4d = PRTree4D(indices, boxes_4d) # 4D boxes# Query with point coordinates
result = tree.query([0.5, 0.5]) # Returns indices
result = tree.query(0.5, 0.5) # Varargs also supported# Insert new rectangle
tree.insert(3, np.array([1.0, 1.0, 2.0, 2.0]))
# Remove rectangle by index
tree.erase(2)
# Rebuild for optimal performance after many updates
tree.rebuild()# Store any picklable Python object with rectangles
tree = PRTree2D()
tree.insert(bb=[0, 0, 1, 1], obj={"name": "Building A", "height": 100})
tree.insert(bb=[2, 2, 3, 3], obj={"name": "Building B", "height": 200})
# Query and retrieve objects
results = tree.query([0.5, 0.5, 2.5, 2.5], return_obj=True)
print(results) # [{'name': 'Building A', 'height': 100}, {'name': 'Building B', 'height': 200}]# Find all pairs of intersecting rectangles
pairs = tree.query_intersections()
print(pairs) # numpy array of shape (n_pairs, 2)
# [[1, 3], [2, 5], ...] # pairs of indices that intersect# Save tree to file
tree.save('spatial_index.bin')
# Load from file
tree = PRTree2D('spatial_index.bin')
# Or load later
tree = PRTree2D()
tree.load('spatial_index.bin')Note: Binary format may change between versions. Rebuild your tree after upgrading.
✅ Good for:
- Large static datasets (millions of boxes)
- Batch queries (parallel processing)
- Spatial indexing, collision detection
- GIS applications, game engines
- Frequent insertions/deletions (rebuild overhead)
- Real-time dynamic scenes with constant updates
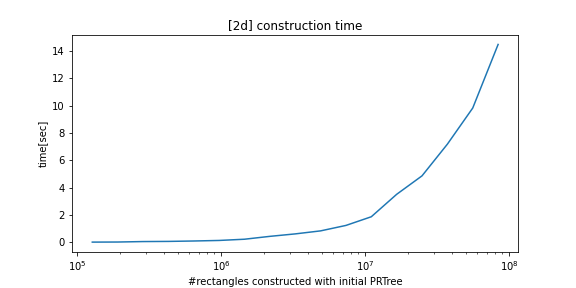
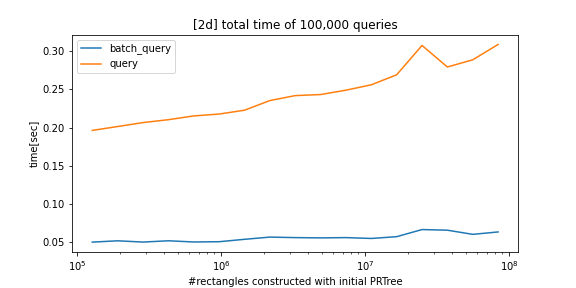
Fast construction and query performance compared to alternatives:
Batch queries use parallel processing for significant speedup.
Boxes must have min ≤ max for each dimension:
# Correct
tree.insert(1, [0, 0, 1, 1]) # xmin=0 < xmax=1, ymin=0 < ymax=1
# Wrong - will raise error
tree.insert(1, [1, 1, 0, 0]) # xmin > xmax, ymin > ymaxAll operations are safe on empty trees:
tree = PRTree2D()
result = tree.query([0, 0, 1, 1]) # Returns []
results = tree.batch_query(queries) # Returns [[], [], ...]The library supports native float32 and float64 precision with automatic selection:
- Float32 input: Creates native float32 tree for maximum speed
- Float64 input: Creates native float64 tree for full double precision
- Auto-detection: Precision automatically selected based on numpy array dtype
- Save/Load: Precision automatically detected when loading from file
The new architecture eliminates the previous float32 tree + refinement approach, providing true native precision at each level for better performance and accuracy.
Read Operations (Thread-Safe):
query()andbatch_query()are thread-safe when used concurrently from multiple threads- Multiple threads can safely perform read operations simultaneously
- No external synchronization needed for concurrent queries
Write Operations (Require Synchronization):
insert(),erase(), andrebuild()modify the tree structure- These operations use internal mutex locks for atomicity
- Important: Do NOT perform write operations concurrently with read operations
- Use external synchronization (locks) to prevent concurrent reads and writes
Recommended Pattern:
import threading
tree = PRTree2D([1, 2], [[0, 0, 1, 1], [2, 2, 3, 3]])
lock = threading.Lock()
# Multiple threads can query safely without locks
def query_worker():
result = tree.query([0.5, 0.5, 1.5, 1.5]) # Safe without lock
# Write operations need external synchronization
def insert_worker(idx, box):
with lock: # Protect against concurrent reads/writes
tree.insert(idx, box)# Clone with submodules
git clone --recursive https://github.com/atksh/python_prtree.git
cd python_prtree
# Install in development mode with all dependencies
pip install -e ".[dev]"For detailed development setup, see DEVELOPMENT.md.
PRTree2D() # Empty tree
PRTree2D(indices, boxes) # With data
PRTree2D(filename) # Load from fileParameters:
indices(optional): Array of integer indices for each bounding boxboxes(optional): Array of bounding boxes (shape: [n, 2*D] where D is dimension)filename(optional): Path to saved tree file
Query Methods:
-
query(*args, return_obj=False)→List[int]orList[Any]- Find all bounding boxes that overlap with the query box or point
- Accepts box coordinates as list/array or varargs (e.g.,
query(x, y)for 2D points) - Set
return_obj=Trueto return associated objects instead of indices
-
batch_query(boxes)→List[List[int]]- Parallel batch queries for multiple query boxes
- Returns a list of result lists, one per query
-
query_intersections()→np.ndarray- Find all pairs of intersecting bounding boxes
- Returns array of shape (n_pairs, 2) containing index pairs
Modification Methods:
-
insert(idx=None, bb=None, obj=None)→None- Add a new bounding box to the tree
idx: Index for the box (auto-assigned if None)bb: Bounding box coordinates (required)obj: Optional Python object to associate with the box
-
erase(idx)→None- Remove a bounding box by index
-
rebuild()→None- Rebuild tree for optimal performance after many updates
Persistence Methods:
-
save(filename)→None- Save tree to binary file
-
load(filename)→None- Load tree from binary file
Object Storage Methods:
-
get_obj(idx)→Any- Retrieve the Python object associated with a bounding box
-
set_obj(idx, obj)→None- Update the Python object associated with a bounding box
Size and Properties:
-
size()→int- Get the number of bounding boxes in the tree
-
len(tree)→int- Same as
size(), allows usinglen(tree)
- Same as
-
n→int(property)- Get the number of bounding boxes (same as
size())
- Get the number of bounding boxes (same as
- Native precision support: True float32/float64 precision throughout the entire stack
- Architectural refactoring: Eliminated idx2exact complexity for simpler, faster code
- Auto-detection: Precision automatically selected based on input dtype and when loading files
- Advanced precision control: Adaptive epsilon, configurable relative/absolute epsilon, subnormal detection
- Fixed critical bug: Boxes with small gaps (<1e-5) incorrectly reported as intersecting
- Breaking: Minimum Python 3.8, serialization format changed
- Added input validation (NaN/Inf rejection)
- Added 4D support
- Object compression
- Improved insert/erase performance
Priority R-Tree: A Practically Efficient and Worst-Case Optimal R-Tree Lars Arge, Mark de Berg, Herman Haverkort, Ke Yi SIGMOD 2004 Paper
- CONTRIBUTING.md - How to contribute to the project
- CHANGES.md - Version history and changelog
- docs/DEVELOPMENT.md - Development environment setup
- docs/ARCHITECTURE.md - Codebase structure and design
- docs/MIGRATION.md - Migration guide between versions
See LICENSE file for details.